doi: 10.56294/mw2024522
ORIGINAL
Investigating Multi-Drug Resistance in MBL-Producing Gram-Negative Bacteria from Clinical Samples: A Phenotypic Approach in Hospitalized Patients
Investigación de la resistencia a múltiples fármacos en bacterias gramnegativas productoras de MBL procedentes de muestras clínicas: Un enfoque fenotípico en pacientes hospitalizados
Naresh Kaushik1
![]() *, Soumya Surath
Panda2
*, Soumya Surath
Panda2 ![]() ,
Nyalam Ramu3
,
Nyalam Ramu3 ![]() ,
Prateek Garg4
,
Prateek Garg4 ![]() ,
Nittin Sharma5
,
Nittin Sharma5 ![]() ,
Uma Bhardwaj6
,
Uma Bhardwaj6 ![]()
1ATLAS SkillTech University, Department of uGDX, Mumbai, Maharashtra, India.
2IMS and SUM Hospital, Siksha ‘O’ Anusandhan (Deemed to be University), Department of Onco-Medicine, Bhubaneswar, Odisha, India.
3Centre for Multidisciplinary Research, Anurag University, Hyderabad, Telangana, India.
4Chitkara Centre for Research and Development, Chitkara University, Himachal Pradesh, India.
5Centre of Research Impact and Outcome, Chitkara University, Rajpura, Punjab, India.
6Noida International University, Department of Biotechnology and Microbiology, Greater Noida, Uttar Pradesh, India.
Cite as: Kaushik N, Panda SS, Ramu N, Garg P, Sharma N, Bhardwaj U. Investigating Multi-Drug Resistance in MBL-Producing Gram-Negative Bacteria from Clinical Samples: A Phenotypic Approach in Hospitalized Patients. Seminars in Medical Writing and Education. 2024; 3:522. https://doi.org/10.56294/mw2024522
Submitted: 26-10-2023 Revised: 28-01-2024 Accepted: 04-05-2024 Published: 05-05-2024
Editor: PhD.
Prof. Estela Morales Peralta ![]()
Corresponding Author: Naresh Kaushik *
ABSTRACT
Introduction: global public health faces a substantial risk due to antimicrobial resistance, especially in clinical settings were tackling the formidable issue of multidrug-resistant Gram-negative bacteria (GNB) is crucial. Metallo-beta-lactamase (MBL)-producing GNB continue to pose a serious threat to medical infection management in hospitals as well as communities.
Method: therapeutic options must be guided by the timely identification of multidrug-resistant (MDR) strains using critical techniques including antimicrobial susceptibility testing (AST) and a MBL monitoring. This study investigates the frequency and transportation as well as antimicrobial sensitivity of GNB that produce MBL and MDR germs in medical specimens were obtained. Aseptically taken from study-eligible in- and out-patients, 491 materials (blood, urine, and wound) produced GNB, which were recovered using conventional bacteriological methods.
Results: to identify the single GNB, determine the AST and discover the GNB that produced MBL. Gram-negative microbes were found to be growing in 130 cases (26,47 %), with the most common isolates being Escherichia coli (15,38 %), Klebsiella pneumoniae (16,92 %), and Burkholderiacepacia (30,76 %). GNB were common in urine samples in other test categories with a frequency of (31,53 %).
Conclusion: to support wise treatment choices, comprehensive antibiotic and MBL screening for drug-resistant GNB is essential. It is highly advised to examine antibiotic usage rules, promote the sensible use of prescription antibiotics, and improve detection of GNB that produces MBL.
Keywords: Gram-Negative Bacteria (GNB); Medical Isolate; Metallo-Beta-Lactamase (MBL); Multidrug-Resistant (MDR).
RESUMEN
Introducción: la salud pública mundial se enfrenta a un riesgo sustancial debido a la resistencia a los antimicrobianos, especialmente en entornos clínicos donde es crucial abordar el formidable problema de las bacterias gramnegativas multirresistentes (GNB). Las GNB productoras de metalo-beta-lactamasas (MBL) siguen representando una grave amenaza para la gestión de las infecciones médicas en hospitales y comunidades.
Método: las opciones terapéuticas deben guiarse por la identificación oportuna de cepas multirresistentes (MDR) mediante técnicas críticas que incluyan pruebas de susceptibilidad antimicrobiana (AST) y un seguimiento de MBL. Este estudio investiga la frecuencia y el transporte, así como la sensibilidad antimicrobiana de GNB que producen MBL y se obtuvieron gérmenes MDR en muestras médicas. Tomadas asépticamente de pacientes hospitalizados y ambulatorios elegibles para el estudio, 491 materiales (sangre, orina y heridas) produjeron GNB, que se recuperaron mediante métodos bacteriológicos convencionales.
Resultados: identificar el único GNB, determinar el AST y descubrir el GNB que produjo MBL. Se observó que en 130 casos (26,47 %) crecían microbios gramnegativos, siendo los aislados más comunes Escherichia coli (15,38 %), Klebsiella pneumoniae (16,92 %) y Burkholderiacepacia (30,76 %). Las GNB fueron comunes en las muestras de orina de otras categorías de pruebas, con una frecuencia del (31,53 %).
Conclusión: para poder elegir un tratamiento adecuado, es esencial realizar un cribado exhaustivo de antibióticos y MBL para detectar GNB resistentes a fármacos. Se recomienda encarecidamente examinar las normas de uso de antibióticos, promover el uso sensato de los antibióticos prescritos y mejorar la detección de GNB que producen MBL.
Palabras clave: Bacterias Gram-Negativas (GNB); Aislado médico; Metalo-Beta-Lactamasa (MBL); Multidrug-Resistant (MDR).
INTRODUCTION
The worldwide public health is facing a dangerous issue due to the establishment and spread of MDR capabilities among GNB that manufacture MBL. The term MDR describes the capacity of these bacteria to resist the actions of many types in antimicrobial medications, making traditional treatment approaches were useless.(1) Possess a greater capacity to hydrolyze a broad range of beta-lactams, involving derivatives of cephalosporins, penicillin’s, and others containing an oxyimino side chain, cephamycin’s, oxepines, and carbapenems.(2)
The generation of these digestive enzymes by pathogenic Gram-negative microorganisms, particularly those in the Enterobacteriaceae group and non-glucose fermenters like Pseudomonas aeruginosa and Acinetobacter baumannii, has significant the clinic and epidemiology implications for the administration of antibiotic radiation therapy and the treatment of Gram-negative infections in healthcare facilities and communities.(3) To its advantageous qualities that include a broad range of antibacterial effects and a strong fatal impact on developing bacteria, beta-lactam antibiotics among the most widely utilized groups of antimicrobial drugs in the medical management of illnesses.(4)
Because beta-lactam antibiotics are widely used, they are inexpensive; non-clinicians could prescribe them incorrectly and people self-medicate resistance to rising and is currently an important threat to public health.(5) Resistance to β-lactam medicines can be acquired by pathogenic bacteria through many ways. These involve decreased permeability, recombination or mutation of transpeptidases that, inhibition by β-lactamases, outflow or porin loss, and diminished porosity. A growing body of research has examined the co-carriage of MDR genes by isolates Gram-negative microorganisms.(6)
The emergence of developed susceptibility to multiple in practice valuable classes of antimicrobial medications, including tetra cyst steroids, trimethoprime, fluoroquinolones, sulfate and polymyxins, to beta-lactic acid. There exist many phenotypic techniques for identifying bacteria that produce MBL. The double-disk synergy examination involving EDTA with Imipenem is the most utilized assays. All of these techniques rely on metal chelators’ capacity to block MBL action, such as Ethylenediamine tetraacetic acid (EDTA) and substances that utilize thiols.(7) The test’s variation is evident in terms of methods, beta-lactam substrates, bacterial species, presence or absence of MBL, inhibitors IMBL and distribution patterns among identified MBL variants in the designated area. A wide variety of clinical examples, including blood, urine and wound specimens obtained from people seeking medical care at three renowned South India health institutions were included in the research.(8)
The key changes in apoptotic and the associated networks that support MDR outlined.(9) Compounds derived from nature that can be employed either alone or in conjunction with traditional therapies. Material regarding to patients or caregivers was gathered using an organized survey.(10) Clinical specimen processing, culture and sensitivity testing, Gram staining, biochemical assays, and the Kirby diffusing disc technique to assess antibiotic susceptibility were then performed, and SPSS version 21 was utilized for analysis. Out of the 153 isolates from various specimens in our investigation, 70 (45,8 %) came from female patients.
Phenotypic and Vitek 2 automated methods were used to identify the etiological the representative and track antibacterial sensitivity in DANI patients. The antibacterial activity of the biosynthesized nanocomposite was also evaluated.(11) Findings: A total of 324 patients received a DANI diagnosis. From DANI patients, a total of 369 microbiological infectious were recovered. Gram-negative bacteria made up most of the pathogenic microorganisms (87 %) and proved opposed to several drugs.
Blood cultures from 1486 patients with probable blood circulation infections between 2018 and 2019 were examined in a cross-sectional investigation.(12) Gram-negative bacteria were categorized using the method of Gram staining, community features, and biological assays; drug-resistant bacteria were tested using antimicrobial susceptibility analysis. Of the gram-negative bacteria, we discovered that 25,7 % produced beta-lactamase and 54,0 % produced extended-spectrum beta-lactam antibiotic-resistance bacteria.
To identify patterns of antibiotic sensitivity and geographic distribution of GNB that produce Metallo- and broad-spectrum beta lactamases evaluated.(13) Our goal was to evaluated the experimental efficacy of minocycline against isolates of Gram-negative bacteria producing beta -lactamase variants found in hospital in Lodz Poland.(14) To examined 102 strains that produced various beta-lactamase, including Klebsiella pneumoniae carbapenem’s (KPC), Metallo-β-lactamase (MBL), oxacillinase (OXA-48) and Guiana extended-spectrum GES.
Acinetobacter baumannii and Pseudomonas aeruginosa were identified in a cross- sectional examination conducted from January to June 2021. Data were gathered through patient database evaluations and interviews, and extended-spectrum β-lactamases (EsβL) and carbapenem’s production were detected.(15) In all, 384 patients took engagement with the research. In all, 30 patients (7,8 %) received good cultivation results for A.
A total of 180 Gram-negative bacterial isolates were examined for antibiotic susceptibility using the disc diffusion method.(16) Among these,40 isolates harbored carbapenems- expressing genes, produced carbapenem’s, and possessed Metallo-beta-lactamases (MBLs). Findings the 40 carbapenem-resistant Gram-negative bacteria (CR-GNB) isolates that were chosen, 28 (70 %) and 20 (50 %) showed phenotypic significant outcomes for the synthesis of MBL and carbapenems, correspondingly.
Structure, innovative entry mode into bacteria and resilience against carbapenem’s contribute to the remarkable potency of Cefetecol against bacterial isolates that demonstrated enhanced resistance or generate carbapenems of all kinds, because of mutations in porin channels and over expression of efflux pumps.(17)
The purpose of this investigation was to describe medical (XDR-PA) isolates that were recovered over a six-month period from three Egyptian hospitals, emphasizing the rising prevalence of hospital-acquired infections.(18) Consequently, a significant proportion of the isolates seventy five percentage were XDR, eighteen percentage were multi-drug resistance (MDR), and only six percentage were non-MDR.
The demography of strains resistant to the discovered mechanisms underlying obstruction, with an emphasis on the generation of variations of β-lactamases, the over expression of β-lactamases, decreased permeability and increased pumps that facilitate efflux.(19)
METHOD
The investigation employed a phenotypic approach to look at MDR in GNB that generate MBL that were separated from hospitalized patients’ clinical samples. The method involved collecting clinical samples from multiple sources in the healthcare setting, then identifying them using defined laboratory techniques to isolate and characterize MBL-causing agents using a holistic phenotypic approach Advanced MDR built-in mechanisms in MDR-positive Gram-negative organisms -Infection -Control- Mechanisms provide important insights
Data collection
The research concentrated on the 11-bed Medical Intensive Care Unit (MICU). The MICU operates as a semi-enclosed room with full-service care staff. The team includes four full-time respiratory therapists, two dedicated emergency room consultants, one ICU-trained nurse serving three a each serving, and a new group of fourteen medical trainees every two months. The care plan included issues with feeding, central venous catheter use, and mechanical ventilation, as well as ICU treatments and events, with organ outcomes including a description of system failure (OSF).
Gram-negative bacteria isolate confirmation using molecular techniques
If an isolate is considered MDR in microbial testing if it has demonstrated resistance to more than two different drugs, this criterion is an important criterion for evaluating the resistance of the isolate a wide range of medications. Researchers use a compound called the MAR index to measure how much resistance a microorganism exhibit. The number of drugs for which the microbial isolate exhibits resistance is divided by the total number for which this index was obtained Essentially, the MAR index provides a statistical picture of the resistance of the isolate against. emphasizing the critical need to actively manage antimicrobial agents and develop new methods for producing resistant bacteria.
Evaluation of mdr and multiple antibiotic resistances (mar) index computation
If an isolate is considered MDR in microbial testing if it has demonstrated resistance to more than two different drugs, this criterion is an important criterion for evaluating the resistance of the isolate a wide range of medications. Researchers use a compound called the MAR index to measure how much resistance a microorganism exhibit. The number of drugs for which the microbial isolate exhibits resistance is divided by the total number for which this index was obtained Essentially, the MAR index provides a statistical picture of the resistance of the isolate against. emphasizing the critical need to actively manage antimicrobial agents and develop new methods for producing resistant bacteria.
Tests for antibiotic susceptibility
Bacterial solutions meeting the requirements for 0,5 McFarland staining were added to Mueller-Hinton agar (MHA) plates, and developed. Several antibiotic discs predetermined on 90 mm petridis using a modified Kirby Bauer disc diffusion method were used for microbial susceptibility studies. Imipenem (10μg), gentamicin (10μg), augmenting (amoxicillin 20μg / clavulanate 10μg), ciprofloxacin (10μg), cefotaxime (30μg), ceftazidime (30μg), aztreonam (30μg), cefepime (30μg), trimethoprim-sulfamethoxazole (2,5 μg), ceftriaxone (30μg), and ofloxacin (5μg) were among the drugs tested. Applying discs to the agar surface facilitated the detection of bacterial sensitivity to different antimicrobial agents. Each disc represented a specific antibody. The efficacy of antibiotics on the studied microorganisms can be determined with reliable and reproducible results, which was promised by this standardized approach
Finding mbl formation using genetic techniques
In the first screening step for potential MBL producers, samples were obtained from GNB that were sensitive to imipenem. The study included a thorough examination of the inhibition sites, paying particular attention to the large ones surrounding imipenem when applied and in combination with ethylenediamine tetra acetic acid EDTA cycles Experiments were conducted for 16 hours to 18 in ventilated atmosphere at 35°C. This pain-related study reveals complex mechanisms of antibiotic sensitivity in these bacterial species and provides important new information on how GNB may be involved in MBL production in the 19th century.
Statistical analysis
The extensive analysis in this study used a robust statistical tool, SPSS software version 25.0. Each variable was tested extensively, which made it possible to examine the complex interactions in the data set more closely. Statistical method for determining p-values and assessing statistical significance of observed correlations Chi-square test was included in the analysis for variability Analysis of factor variability included estimates of odds ratio (OR) and 95 %; confidence interval (CI) about the fundamental level -A critical threshold was established for the value; this phrase indicates that the result is considered significant when it falls below this critical point.
RESULTS AND DISCUSSION
A large 37,88 % of the total incidence distribution was produced by the 186 distinct bacterial isolates that were obtained from the selected research region after an investigation of microbiological samples was conducted. 175 specimens (35,64 %) of this wide range of isolates were effectively identified, providing more information into the complex microbial makeup. 130 of the detected isolates (26,47 % of the total isolates) exhibited traits typical of GNB. The bacterial communities in the studied region are diverse and dynamic, as evidenced by the preponderance of GNB. The understanding of the local microbiological environment is greatly advanced by the rich microbial variety observed in this study, which illuminates the complex distribution patterns and dominant traits. By promoting a greater understanding of the complex ecological dynamics at work in the studied area, these discoveries greatly advance the discipline of microbiology.
The investigation of the frequency distribution is presented in Table 1 which provides an in-depth analysis of GNB isolates obtained from various medical samples. The analytical study presents significant results. Particularly, Escherichia coli are the most common pathogen with an incidence rate of 15,38 %. Klebsiella pneumoniae follows closely with a significant 16,92 % incidence. These results not only provide important new understandings of the microbial ecology. These findings were obtained from the analysis of sample sources, especially with the distribution of Gram-negative isolates, where variance was seen between specimen types. 31,53 % of urine samples included Gram-negative isolates, which was higher than the percentages in both blood (34,61 %) and wound samples (0,76 %). This variation highlights the importance of source-specific variations in the frequencies of bacteria, providing more information into the complex dynamics of bacterial infections at various geographical sites. These findings are crucial in expanding the knowledge of the spatial epidemiology of GNB infections, to provide more information on the variety of GNB. The information provided highlights the necessity of an investigation of bacterial illnesses, taking into the variety of origins and their possible influence on local epidemiological trends.
|
Table 1. Frequency distribution of GNB isolates in clinical samples |
||||
|
Isolates |
Blood sample ( %) |
Wound sample ( %) |
Urine sample ( %) |
Total ( %) |
|
Hafnia alvei (n=5) |
2(40) |
1(20) |
2 (40) |
5 (3,84) |
|
Morganellamorgannii (n=1) |
1 (100) |
0(0) |
0 (0) |
1(0,76) |
|
Acinetobacter baumannii (n=3) |
1 (300) |
1(33,3) |
1 (33,3) |
3 (2,30) |
|
Rhizobium radiobacter (n=4) |
2 (50) |
1(25) |
1(25) |
4 (3,07) |
|
Citrobacter freundi (n=7) |
2 (28,5) |
3(43) |
2 (28,57) |
7 (5,38) |
|
Enterobacter cloaecae complex (n=3) |
1 (33,3) |
0(0) |
2(66,67) |
3 (2,30) |
|
Enterobacter hormaechei (n=6) |
2 (33,3) |
2(33) |
2 (33,33) |
6 (4,61) |
|
Enterobacter kobei (n=4) |
1 (25) |
1(25) |
2 (50) |
4 (3,07) |
|
Enterobacter cloacae cloacae (n=4) |
2 (50) |
1(25) |
1(25) |
4 (3,07) |
|
Enterobacter cloacae dissolvens (n=2) |
0 (0) |
2(100) |
0 (0) |
2 (1,53) |
|
Enterobacter aerogenes (n=7) |
2 (28,57) |
3(43) |
2(28,57) |
7 (5,38) |
|
Pseudomonas aeruginosa (n=8) |
2 (25) |
4(50) |
2 (25) |
8 (6,15) |
|
Proteus mirabilis (n=7) |
2 (28,57) |
3(43) |
2 (28,57) |
7 (5,38) |
|
Burkholderiastabilis (n=2) |
0 (0) |
1(50) |
1 (50) |
2 (1,53) |
|
Burkholderiacepacia (n=13) |
4 (30,76) |
5(38,46) |
4 (30,76) |
13 (30,76) |
|
Klebsiella oxytoca (n=3) |
1 (33,3) |
1(33) |
1 (33,33) |
3 (2,30) |
|
Klebsiella pneumoniae (n=22) |
10 (45,45) |
5(23) |
7 (31,81) |
22 (16,92) |
|
Serratia ficaria (n=2) |
1 (50) |
0(0) |
1 (50) |
2 (1,53) |
|
Serratia fonticola (n=7) |
2 (28,57) |
4 (57,14) |
1 (14,28) |
7 (5,38) |
|
Escherichia coli (n=20) |
7 (35) |
6 (30) |
7 (35) |
20 (15,38) |
|
Total |
45 (34,61) |
44 (0,76) |
41 (31,53) |
130 (100) |
Table 2 presents the details of the Gram-negative bacilli (GNB) species responsible for the distribution of the MBL in the studied area. Of the 130 gram-negative microbial isolates examined, the MBL was 58, accounting for 44,6 % of the total. It is important that every strain of Morganellamorgannii and Hafnia alvei showed a 100 % MBL yield. Other bacteria, including Proteus mirabilis (5,38 % of 7 strains), Rhizobium radiobacter (3,07 % with 4 strains), and Pseudomonas aeruginosa (6,15 % with 8 strains) also exhibited a significant frequency of MBL generation. These results highlight the species diversity of MBL producers found in different GNBs, with specific species exhibiting high levels of MBL production. The data shown in table 2 shed light on the susceptibility of various levels of MBL production to the Gram-negative microbial isolates under investigation, which aids in the detection and resolution of antibiotic resistance in the field in.
|
Table 2. Antibiotic Susceptibility of GNB in the Study Area |
||
|
Isolate |
No. screened |
No. MBL producer ( %) |
|
Enterobacter cloaecae complex |
7 |
12 (171,42) |
|
Klebsiella oxytoca |
8 |
7 (88) |
|
Rhizobium radiobacter |
14 |
4 (28,57) |
|
Enterobacter aerogenes |
8 |
1 (12,5) |
|
E. cloacae dissolvens |
5 |
3 (60) |
|
Acinetobacter baumannii |
19 |
3 (15,78) |
|
Enterobacterhormaechei |
1 |
0 (0) |
|
Morganellamorgannii |
2 |
1 (50) |
|
Enterobacter cloacaecloacae |
3 |
2 (66,67) |
|
Citrobacter freundi |
1 |
3 (300) |
|
Proteus mirabilis |
4 |
1 (25) |
|
Hafnia alvei |
3 |
2 (66,67) |
|
Enterobacter kobei |
7 |
2 (28,57) |
|
Burkelderiastabilis |
25 |
6 (24) |
|
Serratia fonticola |
9 |
3 (33,33) |
|
Pseudomonas aeruginosa |
4 |
4 (100) |
|
Klebsiella pneumoniae |
6 |
2(33,33) |
|
Burkolderiacepacia |
4 |
0 (0) |
|
Total |
130 |
58 (44,6) |
The prevalence of GNB isolates, identified on the basis of the ability to produce MBL in clinical samples, is shown prominently in figure 1. Of the 58 individuals found to produce MBL, the majority, or 51,72 % (30 cases), from urine samples. The important role of water as a reservoir for these antibiotic-resistant bacteria is demonstrated by its high prevalence. The data illustrate an important association between water samples and MBL-producing bacteria, emphasizing the importance of attention and infection prevention measures in this particular clinical setting Understanding the spatial distribution of MBL in samples is important to limit and manage antibiotic resistance in pharmaceutical settings. The results highlight the need for increased awareness and customization, especially when it comes to reducing GNB-induced MBLs and maintaining drug efficacy the emphasis of the. The finding that 17 samples (29,31 %) of MBL-producing isolates were found in wound samples highlights the potential risk posed by wounds as a potential reservoir for the spread of this resistant virus. This study highlights how it need to establish effective infection control measures in wound care settings. The identification of 11 (18,96 %) cases of MBLs in blood indicates the presence of these compounds in blood, emphasizing the need for careful and timely monitoring in medical institutions. These findings have ramifications that go below simple prevalence data, to protect patients’ health, healthcare professionals should keep up with new developments in antibiotic resistance and modify their treatment plans accordingly. As a result, the observations add to continuing the conversation about antibiotic resistance and highlight the urgent need for all-encompassing approaches to handle and control such problems in the medical sector. Table 3 depicts the clinical samples.
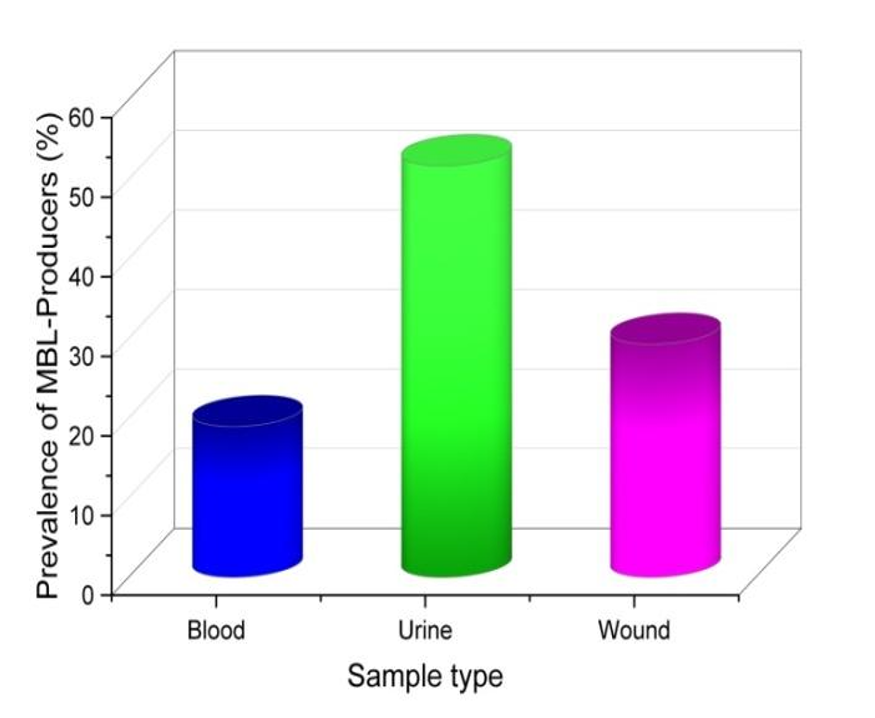
Figure 1. MBL-producing distribution
|
Table 3. Samples with MBL-producing bacteria |
|
|
Sample Type |
Prevalence of MBL-Producers ( %) |
|
Blood |
18,96 |
|
Urine |
51,72 |
|
Wound |
29,31 |
The research carried out in the designated field offers a perceptive examination of the frequency of GNB that are resistant to many drugs, specifically their ability produces MBL it is indicated in table 4. The results highlight a significant pattern, showing that all metallo-beta-lactamase manufacturers (MBL-producers) demonstrated MDR, highlighting the elevated resilience capacities in this subpopulation. The study also reveals that (19,44 %) of people who do not manufacture metallo-beta-lactamase (non-MBL) also showed resistance to many medications, providing information into the wider range of resistant bacteria that exists among people who do not make MBL. These findings highlight the crucial need for focused treatments and monitoring methods to address the escalating threats presented by MDR and gram-negative microbes that provide more information about trends in antibiotic resistance. Table 4 depicts MDR GNB and MBL Production Prevalence.
|
Table 4. MDR and MBL Production Prevalence |
||
|
Physical traits |
No. Of isolates |
No. MDR ( %) |
|
MBL |
58 |
58 (100) |
|
Non-MBL |
72 |
14 (19,44) |
|
Total |
130 |
72 (55,38) |
DISCUSSION
The escalating resistance of GNB to broad-spectrum antibiotics poses a serious public health threat, particularly given the limited availability of antibiotics effective against MBL producers. MBLs zinc-based enzymes responsible for resistance to nearly including carbapenems, underscore the urgency of addressing this issue. Early detection of MBL-producing GNB is crucial to mitigate the spread of multidrug-resistant phenotypes and enhance treatment efficacy. Variability in the proportion of isolated bacteria can arise due to discrepancies in isolation methods and variations in sample collection techniques, thereby introducing potential factors that might exert an impact on bacterial yields.
CONCLUSION
The prevalence of MBL production and multidrug-resistant and GNB reached a significant 44,6 %. Among the isolates, a spectrum of bacteria including E. coli, B. cepacia, H. alvei, E. aerogenes (isolated from urine samples), and K. oxytoca, as well as B. cepacia obtained from blood samples, exhibited the highest degree of resistance known as pan drug resistance (PDR). The alarming finding underscores the urgent need for comprehensive strategies and interventions to address the escalating issue posed by these drug-resistant strains in the healthcare landscape of the region. Antibacterial drugs were met with great resistance by GNB isolates that generated MBLs. It is strongly advised to conduct observation research of these isolates and to use antibiotic sparingly when treating GNB infections that result in MBL. MBL-producing GNB are associated with antibiotic resistance, particularly to β-lactam antibiotics. This poses a significant issue in the treatment of infections caused by these bacteria. The quick and precise identification of bacteria that produce MBL depends on the development of diagnostic methods. This includes the development of molecular and phenotypic assays to enhance detection.
REFERENCES
1. Ejikeugwu C, Nworie O, Saki M, Al-Dahmoshi HO, Al-Khafaji NS, Ezeador C, Nwakaeze E, Eze P, Oni E, Obi C, Iroha I. Metallo-β-lactamase and AmpC genes in Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa isolates from abattoir and poultry origin in Nigeria. BMC microbiology. 2021 Dec;21:1-9. https://doi.org/10.1186/s12866-021-02179-1
2. Noval M, Banoub M, Claeys KC, Heil E. The battle is on: new beta-lactams for the treatment of multidrug-resistant Gram-negative organisms. Current Infectious Disease Reports. 2020 Jan;22:1-9.DOI: https://doi.org/10.1007/s11908-020-0710-9
3. Nordmann P, Poirel L. Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clinical Infectious Diseases. 2019 Nov 13;69(Supplement_7):S521-8. DOI: https://doi.org/10.1093/cid/ciz824
4. Gupta S, Kumar M, Shergill SP, Tandel K. Evaluation of ceftriaxone-sulbactam-disodium edetate adjuvant combination against multi-drug resistant Gram-negative organisms. African Journal of Laboratory Medicine. 2020;9(1):1-6. https://doi.org/10.4102/ajlm.v9i1.991
5. Namaei MH, Yousefi M, Askari P, Roshanravan B, Hashemi A, Rezaei Y. High prevalence of multidrug-resistant non-fermentative Gram-negative bacilli harboring blaIMP-1 and blaVIM-1 metallo-beta-lactamase genes in Birjand, south-east Iran. Iranian journal of microbiology. 2021 Aug;13(4):470. https://doi.org/10.18502/ijm.v13i4.6971
6. Haji SH, Aka ST, Ali FA. Prevalence and characterisation of carbapenemase encoding genes in multidrug-resistant Gram-negative bacilli. PLoS One. 2021 Nov 1;16(11):e0259005. https://doi.org/10.1371/journal.pone.0259005
7. Dumaru R, Baral R, Shrestha LB. Study of biofilm formation and antibiotic resistance pattern of gram-negative Bacilli among the clinical isolates at BPKIHS, Dharan. BMC Research Notes. 2019 Dec;12:1-6. https://doi.org/10.1186/s13104-019-4084-8
8. Emeraud C, Escaut L, Boucly A, Fortineau N, Bonnin RA, Naas T, Dortet L. Aztreonam plus clavulanate, tazobactam, or avibactam for treatment of infections caused by metallo-β-lactamase-producing Gram-negative bacteria. Antimicrobial agents and chemotherapy. 2019 May;63(5):10-128.DOI: https://doi.org/10.1128/aac.00010-19
9. Neophytou CM, Trougakos IP, Erin N, Papageorgis P. Apoptosis deregulation and the development of cancer multi-drug resistance. Cancers. 2021 Aug 28;13(17):4363. DOI: https://doi.org/10.3390/cancers13174363
10. Abd El-Baky RM, Farhan SM, Ibrahim RA, Mahran KM, Hetta HF. Antimicrobial resistance pattern and molecular epidemiology of ESBL and MBL producing Acinetobacter baumannii isolated from hospitals in Minia, Egypt. Alexandria Journal of medicine. 2020 Jan 3;56(1):4-13. https://orcid.org/0000-0001-6094-5653
11. Abbas S, Ejaz H, Jahan S, Younas S, Alzahrani B, Al Farraj DA, Alkufeidy RM. Molecular Detection of blaIMP Genes in Metallo-Beta-Lactamase Producing Clinical Gram-Negative Isolates. Clinical Laboratory. 2019 Aug 1;65(8). 10.7754/Clin.Lab.2019.190202. https://doi.org/10.1080/14656566.2019.1660772
12. Papp-Wallace KM. The latest advances in β-lactam/β-lactamase inhibitor combinations for the treatment of Gram-negative bacterial infections. Expert opinion on pharmacotherapy. 2019 Nov 22;20(17):2169-84.
13. Paskeh MD, Moghaddam MJ, Salehi Z. Prevalence of plasmid-encoded carbapenemases in multi-drug resistant Escherichia coli from patients with urinary tract infection in northern Iran. Iranian Journal of Basic Medical Sciences. 2020 May;23(5):586. https://doi.org/10.22038/ijbms.2020.34563.8199
14. Wang W, Wang X. Prevalence of metallo-β-lactamase genes among Pseudomonas aeruginosa isolated from various clinical samples in China. Journal of Laboratory Medicine. 2020 Sep 25;44(4):197-203. https://doi.org/10.1515/labmed-2019-0162
15. Sarda C, Fazal F, Rello J. Management of ventilator-associated pneumonia (VAP) caused by resistant gram-negative bacteria: which is the best strategy to treat?. Expert Review of Respiratory Medicine. 2019 Aug 3;13(8):787-98. https://doi.org/10.1080/17476348.2019.1632195
16. Kumari M, Verma S, Venkatesh V, Gupta P, Tripathi P, Agarwal A, Siddiqui SS, Arshad Z, Prakash V. Emergence of blaNDM-1 and blaVIM producing Gram-negative bacilli in ventilator-associated pneumonia at AMR Surveillance Regional Reference Laboratory in India. PLoS One. 2021 Sep 8;16(9):e0256308. https://doi.org/10.1371/journal.pone.0256308
17. Yamano Y. In vitro activity of cefiderocol against a broad range of clinically important Gram-negative bacteria. Clinical Infectious Diseases. 2019 Nov 13;69(Supplement_7):S544-51.DOI: https://doi.org/10.1093/cid/ciz827
18. Faujdar SS, Bisht D, Sharma A. Antibacterial activity of Syzygium aromaticum (clove) against uropathogens producing ESBL, MBL, and AmpC beta-lactamase: Are we close to getting a new antibacterial agent?. Journal of family medicine and primary care. 2020 Jan 1;9(1):180-6. 10.4103/jfmpc.jfmpc_908_19
19. Jaloot AS, Owaid MN. Antibiotic Resistance Pattern and Prevalence of Multi-Drug and Extensive Resistant Acinetobacter Baumannii Isolates from Clinical Specimens after Military Operations Western Iraq. Gazi Medical Journal. 2021 Jul 1;32(3). http://dx.doi.org/10.12996/gmj.2021.87
FINANCING
None.
CONFLICT OF INTEREST
None.
AUTHORSHIP CONTRIBUTION
Conceptualization: Naresh Kaushik, Soumya Surath Panda, Nyalam Ramu, Prateek Garg, Nittin Sharma, Uma Bhardwaj.
Research: Naresh Kaushik, Soumya Surath Panda, Nyalam Ramu, Prateek Garg, Nittin Sharma, Uma Bhardwaj.
Writing - original draft: Naresh Kaushik, Soumya Surath Panda, Nyalam Ramu, Prateek Garg, Nittin Sharma, Uma Bhardwaj.
Writing – proofreading and editing: Naresh Kaushik, Soumya Surath Panda, Nyalam Ramu, Prateek Garg, Nittin Sharma, Uma Bhardwaj.