doi: 10.56294/mw2024535
ORIGINAL
Current Approaches and Clinical Implications of Epigenetic Modifications in Cancer Therapy
Enfoques actuales e implicaciones clínicas de las modificaciones epigenéticas en la terapia del cáncer
Kukatla
Tejesh1 ![]() *,
Lovish Dhingra2
*,
Lovish Dhingra2 ![]() , Vinima Gambhir3
, Vinima Gambhir3 ![]() , Amit Kumar4
, Amit Kumar4 ![]() , Nibedita Sahoo5
, Nibedita Sahoo5 ![]() , Manti Debnath6
, Manti Debnath6 ![]() , Malathi.H7
, Malathi.H7 ![]()
1Centre for Multidisciplinary Research, Anurag University. Hyderabad, Telangana, India.
2Chitkara Centre for Research and Development, Chitkara University. Himachal Pradesh, India.
3Department of ISME, ATLAS SkillTech University. Mumbai, Maharashtra, India.
4Centre of Research Impact and Outcome, Chitkara University. Rajpura, Punjab, India.
5Department of Pathology, IMS and SUM Hospital, Siksha ‘O’ Anusandhan (Deemed to be University). Bhubaneswar, Odisha, India.
6School of Allied Health Sciences, Noida International University. Greater Noida, Uttar Pradesh, India.
7Biotechnology and Genetics, JAIN (Deemed-to-be University). Bangalore, Karnataka, India.
Cite as: Tejesh K, Dhingra L, Gambhir V, Kumar A, Sahoo N, Debnath M, et al. Current Approaches and Clinical Implications of Epigenetic Modifications in Cancer Therapy. Seminars in Medical Writing and Education. 2024; 3:535. https://doi.org/10.56294/mw2024535
Submitted: 11-11-2023 Revised: 27-02-2024 Accepted: 20-06-2024 Published: 21-06-2024
Editor: PhD.
Prof. Estela Morales Peralta ![]()
Corresponding Author: Kukatla Tejesh *
ABSTRACT
Epigenetic changes are very important for the growth and spread of cancer because they control gene expression without changing the DNA sequence. Some of these changes, like DNA methylation, histone modifications, and non-coding RNA control, can turn on oncogenes, turn off tumor suppressor genes, and make cancer cells more aggressive. Because epigenetic changes can be undone, they are considered potential therapeutic targets for treating cancer. This essay talks about the latest methods to epigenetic medicines and what they mean for cancer care. The main goal of epigenetic treatments is to undo the abnormal epigenetic changes that cause cancer to spread. Some of the epigenetic drugs that have been studied the most are DNA methyltransferase inhibitors (like azacytidine) and histone deacetylase inhibitors (like vorinostat). Several of these drugs are already in use or are going through clinical studies. Besides that, going after non-coding RNAs like microRNAs and long non-coding RNAs has become an interesting way to change gene expression in cancer cells. Using epigenetic medicines along with common treatments like chemotherapy, immunotherapy, and targeted therapy has been shown to have better healing benefits, sometimes leading to better patient results. Epigenetic changes have a huge range of clinical effects. Epigenetic analysis of cancers can give useful signs for figuring out prognosis and treatment reaction, which makes it possible to create individual treatment plans. Also, epigenetic treatments might be able to beat drug resistance by going after the epigenetic processes that make traditional medicines less effective. However, there are problems with using epigenetic treatments in the field, such as making sure the drugs are safe, making sure they are bioavailable, and finding good ways to give the drugs.
Keywords: Epigenetics; Cancer Therapy; DNA Methylation; Histone Modifications; Personalized Treatment.
RESUMEN
Los cambios epigenéticos son muy importantes para el crecimiento y la propagación del cáncer porque controlan la expresión de los genes sin cambiar la secuencia del ADN. Algunos de estos cambios, como la metilación del ADN, las modificaciones de las histonas y el control del ARN no codificante, pueden activar oncogenes, desactivar genes supresores de tumores y hacer que las células cancerosas sean más agresivas. Dado que los cambios epigenéticos pueden deshacerse, se consideran posibles dianas terapéuticas para tratar el cáncer. Este ensayo habla de los últimos métodos de los medicamentos epigenéticos y lo que significan para el tratamiento del cáncer. El principal objetivo de los tratamientos epigenéticos es deshacer los cambios epigenéticos anormales que provocan la propagación del cáncer. Algunos de los fármacos epigenéticos más estudiados son los inhibidores de la ADN metiltransferasa (como la azacitidina) y los inhibidores de la histona deacetilasa (como el vorinostat). Varios de estos fármacos ya se utilizan o están siendo objeto de estudios clínicos. Además, la búsqueda de ARN no codificantes como los microARN y los ARN no codificantes largos se ha convertido en una forma interesante de modificar la expresión génica en las células cancerosas. Se ha demostrado que el uso de medicamentos epigenéticos junto con tratamientos comunes como la quimioterapia, la inmunoterapia y la terapia dirigida tiene mejores beneficios curativos, lo que a veces se traduce en mejores resultados para los pacientes. Los cambios epigenéticos tienen una enorme variedad de efectos clínicos. El análisis epigenético de los cánceres puede dar señales útiles para conocer el pronóstico y la reacción al tratamiento, lo que permite crear planes de tratamiento individuales. Además, los tratamientos epigenéticos podrían vencer la resistencia a los fármacos actuando sobre los procesos epigenéticos que restan eficacia a los medicamentos tradicionales. Sin embargo, el uso de tratamientos epigenéticos sobre el terreno plantea problemas, como asegurarse de que los fármacos son seguros, garantizar su biodisponibilidad y encontrar buenas formas de administrarlos.
Palabras clave: Epigenética Terapia del Cáncer; Metilación del ADN; Modificaciones de las Histonas; Tratamiento Personalizado.
INTRODUCTION
Cancer is a complicated disease with many causes. It is marked by excessive cell growth, multiplication, and spread. A mix of genetic defects and epigenetic changes cause cancer to grow. These changes help tumors start, spread, and metastasize. For decades, genetic changes like point mutations, insertions, and deletions have been the main focus of cancer study. However, recent progress has shown how important epigenetics is to cancer biology. Epigenetic modifications, which are changes in gene expression that are passed down but don't involve changes in the DNA code itself, have become important in controlling the growth of cancer. By changing the shape and accessibility of chromatin and DNA without changing the genetic code, these changes can affect gene translation. DNA methylation, histone modifications, and non-coding RNA control are the three types of epigenetic changes that are most often linked to cancer.(1) Adding a methyl group to the 5' position of cytosine residues in CpG dinucleotides is what DNA methylation is all about. This silences genes. In cancer, abnormal DNA methylation patterns can turn off genes that stop tumors from growing, which helps new tumors grow. Changes to histones, like acetylation, methylation, phosphorylation, and ubiquitination, control the structure of chromatin and how DNA can be accessed by the machinery for transcription. These changes can either make gene expression go up or down, and changes in the patterns of histone modifications are often seen in many types of cancer. Cancer has also been linked to non-coding RNAs, especially microRNAs and long non-coding RNAs.(2)
These molecules control gene translation after transcription. They can change the activity of oncogenes and tumor suppressor genes, which affects how tumors grow and spread. Because epigenetic changes can be undone, they are good targets for therapeutic action. Genetic defects are permanent, but epigenetic changes can be changed or "reset," which means that the faulty gene expression patterns that are linked to cancer might be able to be fixed or reversed. Because of this, epigenetic treatments have gotten a lot of attention in the study of cancer. The goal of these treatments is to target and change the epigenetic environment of cancer cells. They do this by either turning on tumor suppressor genes that have been turned off or stopping the expression of genes that help tumors grow.(3) A number of epigenetic drugs have already been made and tried in hospitals, with different levels of success. There are two types of epigenetic drugs that have shown potential in cancer treatment.
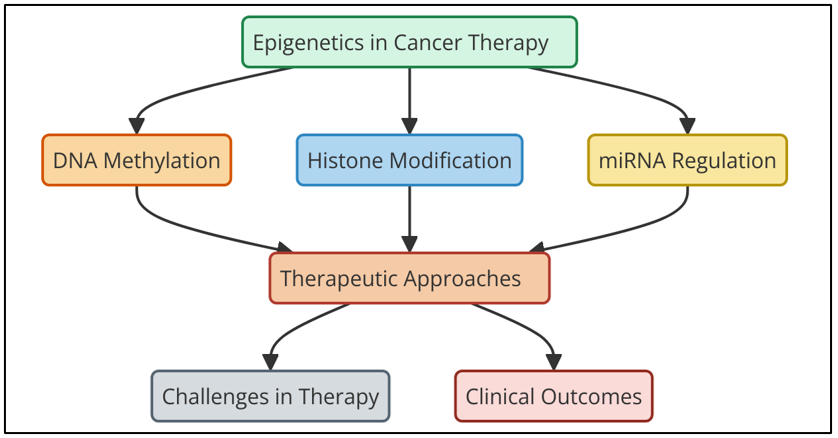
Figure 1. Current Approaches and Clinical Implications of Epigenetic Modifications in Cancer Therapy
Figure 1 represents the current approaches and clinical implications of epigenetic modifications in cancer therapy. It highlights strategies such as DNA methylation inhibitors, histone modification regulators, and non-coding RNA targeting. These approaches modulate gene expression, offering precision treatments that enhance efficacy, reduce resistance, and improve patient outcomes, underscoring their significance in personalized oncology. These are DNA methyltransferase inhibitors (DNMTis) and histone deacetylase inhibitors (HDACis). DNMTis, like azacytidine and decitabine, stop DNA methyltransferases from doing their work. DNA methyltransferases are the chemicals that include methyl groups to DNA. These medicines can turn on tumor suppressor genes that have been turned off, making them work once more and possibly ceasing the tumor from developing. Histone deacetylases are proteins that take acetyl bunches off of histones. HDACis, like vorinostat and romidepsin, halt these proteins from working. By blocking HDACs, these drugs make chromatin more open, which increments the expression of qualities that halt tumors from developing and diminishes the expression of qualities that cause cancer. Beside these well-known medicines, there's developing intrigued in blending epigenetic drugs with other sorts of treatment, like immunotherapy, chemotherapy, and focused on treatment.(4) The thought behind combination medications is to utilize the way that epigenetic changes work with the way that regular cancer drugs work to induce way better comes about. For illustration, epigenetic medications may make cancer cells more touchy to chemotherapy or make immunotherapies work way better by turning on qualities that decrease tumors and making it less demanding for the immune system to discover tumor cells. Indeed in spite of the fact that epigenetic medications appear a lot of promise, they are difficult to utilize in genuine life.
Related work
In recent years, the study of epigenetics in cancer medicines has developed a lot. Researchers are now trying to figure out how changes in epigenetics cause cancer to spread and how to reverse these changes to help with treatment. A lot of research has been done on different epigenetic changes—mainly DNA methylation, histone modifications, and non-coding RNA regulation—and how they affect the growth of tumors, as well as the possibility of treating these changes in cancer treatment. DNA methylation has been the subject of some of the oldest and most important studies. Cancer cells always have DNA methylation patterns that aren't normal. For example, tumor suppressor genes are often over-methylated while oncogenes are under-methylated.(5) One well-known epigenetic change in many types of cancer is the hypermethylation of the p16INK4a gene, which stops the gene from working and encourages uncontrolled cell growth. A lot of research, like that by Baylin and Jones (2011), has looked at the role of DNA methylation in cancer and the treatment promise of DNA methyltransferase inhibitors (DNMTis), like decitabine and azacytidine. In clinical studies, these inhibitors have shown potential by turning on tumor suppressor genes that were turned off and making patient results better, especially in blood cancers.(6)
At the same time, study on changes to histones has shown how important they are for controlling gene translation. It has been found that histone acetylation and methylation can change how key oncogenes and tumor suppressor genes are expressed. For instance, blocking histone deacetylases (HDACs) with drugs like vorinostat has been shown to help treat some types of cancer, like cutaneous T-cell lymphoma. Marks and Breslow (2007) wrote a study that talked about the treatment possibilities of HDAC inhibitors as well as the problems that come up when they are used in real life, like side effects and drug tolerance.(7) Researchers are looking into other changes that can be made to histones, like histone methylation, to see if they can change the chromatin environment and reset cancer cells. Cancer study has also paid a lot of attention to non-coding RNAs, such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs). By attaching to mRNA and stopping translation, miRNAs are very important for controlling gene expression. Misregulation of miRNAs has been linked to the growth of several types of cancer, such as lung, breast, and prostate cancer.
|
Table 1. Summary of Related work |
|||
|
Method |
Approach |
Limitation |
Benefits |
|
DNA Methyltransferase Inhibitors (DNMTis) |
Inhibition of DNA methyltransferases to reverse gene silencing in cancer. |
Potential for off-target effects and toxicity. |
Potential to reverse epigenetic silencing of tumor suppressor genes. |
|
Histone Deacetylase Inhibitors (HDACis)(8) |
Inhibition of histone deacetylases to restore gene expression and modify chromatin. |
Limited specificity and potential side effects, such as hematological toxicities. |
Can restore gene expression and inhibit oncogene activation. |
|
Bromodomain Inhibitors |
Targeting bromodomain-containing proteins to inhibit oncogene expression. |
Challenges in drug delivery and specificity for tumor cells. |
Provides a new mechanism to block tumor-promoting proteins. |
|
CRISPR-based Epigenetic Editing(9) |
Use of CRISPR/Cas9-based systems to modify epigenetic marks directly. |
Ethical concerns, delivery challenges, and potential off-target effects. |
Allows precise modification of the epigenome at specific sites. |
|
Non-coding RNA Modulation |
Modulation of miRNAs or lncRNAs to regulate cancer-related genes. |
Limited understanding of non-coding RNA functions in cancer biology. |
Can restore normal gene expression and inhibit oncogenic pathways. |
|
Combination Therapy (Epigenetic + Chemotherapy) |
Combining epigenetic inhibitors with chemotherapy to enhance treatment efficacy. |
Increased side effects due to chemotherapy, potential for drug resistance. |
Synergistic effects with chemotherapy, improving efficacy. |
|
Combination Therapy (Epigenetic + Immunotherapy)(10) |
Combining epigenetic inhibitors with immunotherapy to improve immune response. |
Higher toxicity rates, especially in combination with immunotherapy. |
Improved immune response and tumor-specific targeting with immunotherapy. |
|
Epigenetic Targeting of Tumor Microenvironment |
Targeting the epigenetic regulation of the tumor microenvironment to enhance therapy. |
Tumor microenvironment complexity, making targeted therapy challenging. |
May enhance efficacy of existing therapies by modifying the tumor microenvironment. |
|
Synthetic Lethality in Epigenetic Therapy |
Exploiting synthetic lethal interactions in cancer cells with altered epigenetic states. |
Difficulty in identifying and exploiting specific synthetic lethal interactions. |
Identifying vulnerabilities in cancer cells with altered epigenetic patterns. |
|
Epigenetic Reprogramming via Small Molecules |
Using small molecules to induce epigenetic changes that reprogram cancer cells. |
Challenges in achieving consistent and long-lasting reprogramming effects. |
Promising for reprogramming tumor cells back to normal, reducing malignancy. |
|
Gene Editing for Epigenetic Modifications(11) |
Direct gene editing to modify epigenetic changes at specific loci in the genome. |
Technical challenges in precise gene editing without unintended consequences. |
Offers precision in targeting specific epigenetic changes with minimal off-target effects. |
|
RNA Interference Targeting Epigenetic Pathways |
Using RNA interference to suppress epigenetic regulators or signaling pathways in cancer. |
Off-target effects and delivery challenges for RNA-based therapies. |
Can reduce the expression of specific epigenetic regulators and restore tumor suppressors. |
Understanding epigenetics in cancer
Definition and key concepts of epigenetics
If you want to learn more about epigenetics, you can look into changes that can be passed down through genes that don't affect the DNA code. Genetic mutations change the order of the DNA's nucleotides. Epigenetic modifications change the structure of DNA or proteins that are linked to DNA to control gene function. These changes don't change the genetic code itself; instead, they change how genes are produced by either turning them on or off. Epigenetic changes are changeable and can sometimes be undone because they are affected by things in the environment, the decisions we make in our daily lives, and getting older. DNA methylation is one of the main ways that epigenetics controls genes. It involves adding a methyl group (CH₀) to the 5' carbon of cytosine residues, which are usually found in CpG dinucleotides. DNA methylation often turns off genes by stopping transcription factors from joining or drawing in proteins that stop transcription. When it comes to cancer, DNA methylation patterns that aren't normal can help the disease get worse.(12) Hypermethylation of tumor suppressor genes, for instance, can stop them from working, while hypomethylation of oncogenes can turn them on, which can help the tumor grow. Modifying histones is another important process. Histones are proteins that help DNA fit into chromatin, which is a very small structure. Changing histones chemically in ways like acetylation, methylation, and phosphorylation affects the shape of chromatin and gene activation. For example, histone acetylation usually makes genes more active by easing up the structure of chromatin, while methylation can either make genes more active or less active, based on the situation.(13) Because of these changes, the cell can change how genes are expressed in reaction to different messages from inside and outside the cell.
Mechanisms of epigenetic modifications
Epigenetic changes are ways to control gene activity without changing the DNA code itself. Chemical changes are made to DNA, histones (proteins that wrap DNA around them), and non-coding RNA molecules as part of these changes. These changes affect the shape of chromatin, which controls gene expression by making it easier or harder for transcription to happen. DNA methylation, histone modification, and control by non-coding RNAs are the major ways that epigenetic changes happen. One of the epigenetic changes that has been studied the most is DNA methylation. It includes adding a methyl group (-CH₀) to the 5' end of cytosine residues, which usually happens in CpG dinucleotides. DNA methylation typically turns off genes by stopping transcription factors from joining or by bringing in methyl-binding proteins that make chromatin denser, which makes it harder for transcription to happen. Cancer often has DNA methylation patterns that aren't working right.(14) For example, tumor suppressor genes may be over-methylated, which stops them from working, or oncogenes may be under-methylated, which can turn them on and help the tumor grow. Histone Modifications are when chemical groups (acetyl, methyl, phosphate, etc.) are added to or taken away from histone proteins, which are important parts of chromatin. These changes can make the chromatin structure looser or tighter, which can affect how genes are expressed. Histone acetylation, for example, removes the positive charge from histones so they don't connect as much with the negatively charged DNA. This makes the chromatin structure more open and turns on genes. On the other hand, based on the type of histone methylation and where it is located on the histone tails, it can either turn genes off or on.(15)
Epigenetic alterations in cancer
Alterations in epigenetics are very important for how cancer starts, grows, and spreads. Although these changes don't involve changing the DNA code itself, they can make genes turn on or off in the wrong way, which can help normal cells turn cancerous. DNA methylation, histone modifications, and non-coding RNA instability are the main epigenetic changes seen in cancer. DNA methylation is one of the epigenetic changes that we know the most about in cancer. Cancer cells often have DNA methylation patterns that aren't normal. For example, tumor suppressor genes may be hypermethylated while oncogenes may be hypomethylated. The promoter regions of tumor suppressor genes are often not modified in normal cells, which lets the genes be expressed.(16) But in cancer cells, hypermethylation of these promoter regions turns off these important genes, which usually stop cells from growing out of control. Hypomethylation of oncogenes or genomic areas, on the other hand, can make them active, which can help tumors grow. Changes to histones are also very important in the growth of cancer. Histones are proteins that wrap DNA up into chromatin. Changing their chemical makeup can have a big effect on how genes are expressed. Histone acetylation and methylation patterns that aren't usual are often seen in cancer. For instance, less histone acetylation makes chromatin structure denser, which lowers gene expression.
Related algorithms in epigenetic cancer research
Machine Learning and Data Mining Approaches
In epigenetic cancer investigate, machine learning (ML) and information mining have become exceptionally useful apparatuses for analyzing huge, complicated datasets that come from genome and epigenomic considers. These strategies are exceptionally imperative for finding designs of epigenetic changes that cause cancer to develop and spread. It can use machine learning strategies to see at information around DNA methylation, histone change, and non-coding RNA expression. This may assist you discover conceivable signals for early identification, expectation, and treatment reaction. Back vector machines (SVMs), random forests, and neural systems are a few of the directed learning strategies that are frequently utilized to partition cancer sorts into bunches based on epigenetic designs. These models learn from labeled information to figure how cancer will turn out or how pharmaceutical will work. By looking at designs of epigenetic changes, unsupervised learning strategies like k-means and hierarchical grouping can show mystery subgroups inside distinctive sorts of cancer.(17) Information mining can moreover offer assistance find links between changes in epigenetics and quality expression or mutations, which can offer assistance us get it how cancer works. Machine learning models can give us a better understanding of how cancer cells work by combining different types of genomic information, like DNA methylation, gene expression, and copy number changes. By speculating which epigenetic changes are most likely to react to certain drugs, these methods may lead to more personalized treatments.
Algorithms for predicting epigenetic alterations in cancer
It is hard to make predictions about epigenetic changes that happen in cancer since the data is so complicated and has a lot of dimensions. A number of computer and machine learning methods have been made to guess these changes. They are ordinarily based on multi-omics information, which incorporates DNA methylation, histone adjustments, and non-coding RNA expression. These strategies help find the epigenetic changes that cause tumors to develop, which helps specialists analyze cancer way better and make decisions approximately treatment. An often-used strategy is the Back Vector Machine (SVM). It is very great at isolating different types of cancer based on epigenetic designs. In a put with numerous measurements, SVMs discover a hyperplane that best isolates distinctive sorts of information into bunches. In epigenetics, SVMs can be instructed to utilize DNA methylation or histone alter characteristics to figure the type of cancer an individual has or how well they will do after treatment.(18) Random Forests is another solid strategy. It is an outfit learning method that can work with big, complicated datasets.
Step 1: Data Preprocessing and Feature Extraction
The first step involves data preprocessing and feature extraction. This includes the cleaning of raw epigenetic data (e.g., DNA methylation levels, histone modifications, and non-coding RNA expression) and extracting relevant features that can be used for prediction. This step also includes normalization and handling missing values.
Mathematical Equation for Data Normalization:
![]()
Step 2: Model Selection and Training
Once the data is preprocessed, an appropriate machine learning model (e.g., support vector machine (SVM), decision trees, or neural networks) is selected and trained using the dataset. The goal here is to learn the relationship between the input features (epigenetic alterations) and the target variable (cancer type, patient outcome, etc.).
Mathematical Equation for SVM Model:
![]()
Where:
w is the weight vector.
x is the input feature vector.
b is the bias term.
f(x) is the decision function.
Step 3: Model Evaluation and Validation
After training the model, it is evaluated using performance metrics like accuracy, precision, recall, and F1-score. Cross-validation is often used to assess the model's generalization ability.
Mathematical Equation for Accuracy:
![]()
Where:
TP = True Positives.
TN = True Negatives.
FP = False Positives.
FN = False Negatives.
Step 4: Prediction and Biomarker Identification
Finally, the trained model is used to make predictions on new, unseen epigenetic data. If the model identifies patterns related to specific biomarkers (e.g., methylation sites or histone marks), these biomarkers can be considered for further validation and potential clinical application.
Mathematical Equation for Prediction:
![]()
Classification models for identifying epigenetic biomarkers
It is exceptionally important to utilize classification models to discover epigenetic biomarkers that can help with identifying, prognosticating, and predicting how cancer will react to treatment. These models utilize diverse sorts of information, like DNA methylation, histone changes, and non-coding RNA expression levels, to sort cancer tests into diverse groups or guess how patients will do. These models can discover biomarkers that might not be able to be found using standard genome strategies by looking at epigenetic marks. This gives us new information about how cancer works. Support Vector Machines (SVMs) are exceptionally well known classification models since they can bargain with expansive sums of information and discover non-linear associations between epigenetic markers. SVMs have been utilized in cancer ponders to sort distinctive sorts of cancer into bunches based on DNA methylation levels or histone alter patterns. SVMs discover the best hyperplane that divides groups, which makes sure that precise forecasts of cancer subtypes are made. Based on epigenetic markers, this demonstrate has been particularly accommodating for telling the contrast between cancerous and non-cancerous cells or speculating how long a persistent will live. Irregular Timberlands is another popular classification strategy.(19) It is an outfit learning procedure that employments different choice trees to create forecasts more exact. Arbitrary Timberlands are extraordinary for managing with huge datasets since they can appear how diverse DNA characteristics associated with each other. Random Forests have been utilized in cancer studies to discover vital DNA methylation locales or histone marks that are connected to certain sorts of cancer or sedate resistance.
Epigenetic Signature Analysis Algorithms
Understanding the complicated control components that administer quality expression and finding conceivable signs for cancer discovery, expectation, and treatment reaction are made conceivable by epigenetic signature examination strategies. These systems see at epigenetic data, like DNA methylation, histone changes, and non-coding RNA expression levels, to discover designs that appear how typical cells are distinctive from unusual cells or how cancer bunches and understanding comes about will turn out. Differential Methylation research may be a strategy that's frequently utilized for epigenetic marker investigate.(20) To discover differentially checked districts (DMRs), this strategy compares the DNA methylation designs of sound and cancerous cells. Many types of statistical tests, counting t-tests, ANOVA, and more advanced ones like the Mann-Whitney U test or limma (straight models for microarray information), are utilized to find areas where methylation changes are important. These DMRs can be used as biomarkers to find cancer or to show how the growth is getting worse.(21)
Step 1: Data Preprocessing and Feature Selection
The first step involves preprocessing the raw epigenetic data (e.g., DNA methylation, histone modifications, non-coding RNA expression) and selecting the most relevant features that contribute to the variation in the dataset. This includes filtering noisy data, handling missing values, and performing normalization.
Mathematical Equation for Data Normalization:
![]()
Where:
X{ij} is the raw data value for feature j in sample i.
μj is the mean of feature j.
σj is the standard deviation of feature j.
X'{ij} is the normalized value for feature j in sample i.
Step 2: Clustering and Signature Extraction
In this step, clustering algorithms like k-means or hierarchical clustering are used to group similar epigenetic profiles. The goal is to identify distinct groups or clusters that share common epigenetic alterations. The signature of each cluster is extracted to define the epigenetic profile for each group.
Mathematical Equation for k-means Clustering:
![]()
Where:
n is the number of samples.
K is the number of clusters.
r{ik} is the responsibility of cluster k for sample i.
Step 3: Signature Validation and Identification of Biomarkers
After extracting the epigenetic signatures, they are validated using statistical tests and machine learning models to identify potential biomarkers associated with cancer types or outcomes. The identified biomarkers can then be subjected to further validation in clinical settings.
Mathematical Equation for Signature Validation (e.g., using t-test):
![]()
Where:
μ1 and μ2 are the means of the two groups being compared.
σ12 and σ22 are the variances of the two groups.
n1 and n2 are the sample sizes of the two groups.
Current epigenetic therapies in cancer treatment
Epigenetic inhibitors: types and mechanisms
It is a nucleoside analog that is added to DNA when it copies itself. Once it is added, azacytidine stops DNMTs from methylating DNA by forming a chemical bond with the enzyme. So, azacytidine lowers the amount of methyl groups in the genome. This turns on tumor suppressor genes that had been turned off, which may help cells work normally again. This restart can cause apoptosis (programmed cell death) in tumor cells, which stops the growth of the tumor. By causing partial differentiation and returning normal blood cell production, azacytidine has shown promise in the treatment of hematopoietic cancers like myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML).
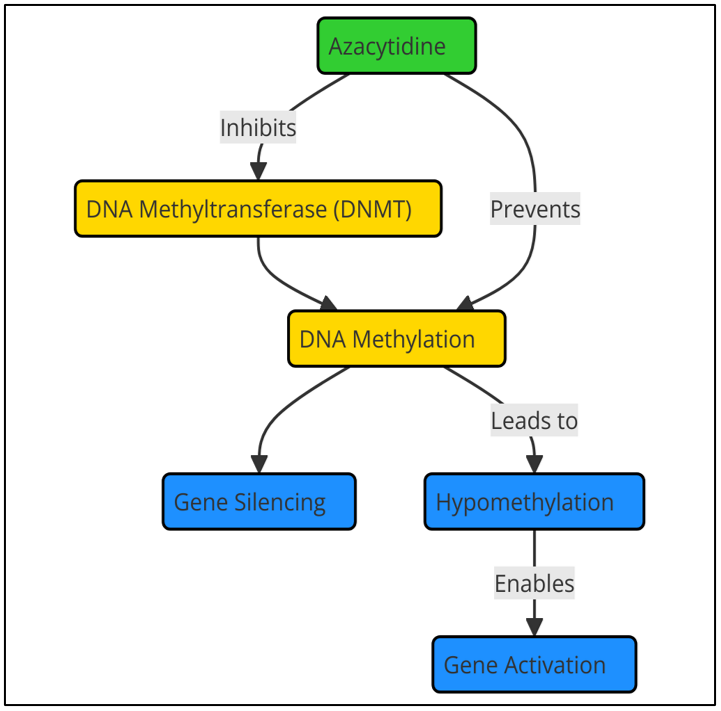
Figure 2. Action of DNA methyltransferase inhibitors like azacytidine
Even though they work, DNMTs like azacytidine have some problems. Gene expression changes caused by the drug are often slow and may need long-term care. Responses vary from patient to patient. Also, taking azacytidine can cause side effects like sickness, tiredness, and a decrease in bone marrow function. To get around these problems, researchers are looking into combination medicine, workflow illustrate in figure 2. In these, DNMTis are used with other treatments, like immunotherapy or chemotherapy, to make them work better and reduce resistance.
Histone deacetylase inhibitors (e.g., vorinostat)
Histone deacetylase inhibitors (HDACis) are a type of epigenetic treatments that work by targeting histone deacetylases (HDACs), which are enzymes that take acetyl groups off of histones. Histones are proteins that DNA wraps around to form chromatin. Acetylation of histones is linked to an open chromatin structure that helps genes get translated. On the other hand, deacetylation makes chromatin more crowded, which stops genes from expressing themselves. A lot of cancers have HDACs that work too much, which turns off tumor suppressor genes and turns on oncogenes. HDACis work by stopping these enzymes from doing their jobs.
Targeting non-coding RNAs for therapy
MicroRNAs
MicroRNAs, or miRNAs, are short RNA molecules that don't code for anything. They are usually between 20 and 24 bases long and are very important for controlling gene activation after transcription. They do their job by attaching to the 3' untranslated region (UTR) of specific mRNA molecules. This either breaks down the mRNA or stops it from being translated into a protein. An enormous number of biological processes are controlled by miRNAs, such as cell cycle control, death, growth, and defense reactions. Because they play a big part in controlling genes, miRNAs have become very important in cancer biology, where their abundance is often changed. Depending on the genes they target, miRNAs can either help cancer grow (oncomiRs) or stop it from growing (suppressors). Some miRNAs are increased in cancer cells, which either stops tumor-suppressor genes from working or helps cells grow, migrate, and fight cell death. On the other hand, when some miRNAs are downregulated or lost, they can cause oncogenes to become overexpressed, which helps cancer spread. For instance, miR-21 is often increased in breast, lung, and colon cancer, among others. It helps tumors grow and spread by targeting genes that stop tumors from spreading, such as PTEN and PDCD4. MiRNAs are good targets for therapy because they play a key role in cancer. MiRNA mimics are man-made molecules that are meant to restore the production of miRNAs that reduce tumors but are downregulated in cancer cells. On the other hand, molecules called antagomiRs or miRNA inhibitors are made to stop overexpressed oncomiRs from doing their bad things to gene expression. For example, the known tumor inhibitor miR-34a is often downregulated in cancers. Increasing its transcript has shown promise in animal models.
Future directions and emerging trends
New epigenetic targets in cancer therapy
New discoveries in cancer research have led to the discovery of several potential new epigenetic targets that can be used for therapy. These are different from older methods like DNA methyltransferase inhibitors (DNMTis) and histone deacetylase inhibitors (HDACis). The goal of these new targets is to undo the epigenetic changes that cause cancer to spread by focusing on different levels of gene control, such as chromatin remodeling, non-coding RNA modulation, and histone modifications. Bromodomain and extraterminal domain (BET) proteins are a new target. These proteins help recognize acetylated lysine residues on histones. BET proteins, like BRD4, are very important for controlling gene expression because they bring transcriptional machinery to chromatin. It has been suggested that BRD4 helps oncogenes like MYC become more active in cancer. In preclinical studies, BET inhibitors like JQ1 have shown promise. They work by messing up the function of BRD4, which lowers MYC levels and stops tumor growth, especially in cancers like leukemia, breast cancer, and small cell lung cancer. Another new epigenetic target is SWI/SNF chromatin modification complexes. These complexes change the shape of chromatin and make it easier for genes to release themselves. Cancers often have changes to these complexes, which makes chromatin modification not work right and changes gene expression. Researchers are working on ways to target certain parts of the SWI/SNF complex, like SMARCA4 and ARID1A.
Advances in CRISPR-based epigenetic editing
CRISPR-based epigenetic editing is an area that is growing quickly. It uses the accuracy of the CRISPR/Cas9 system to change the epigenome directly, which opens up new ways to treat cancer. On the other hand, joining dCas9 with histone deacetylases (HDACs) or histone methyltransferases (HMTs) can turn off genes or make certain tumor suppressor genes more active. These new discoveries make it possible to precisely change gene expression at both the transcriptional and post-transcriptional levels. This lets researchers study how epigenetic changes in cancer affect how genes work.
RESULTS AND DISCUSSION
It looks like new developments in epigenetic treatments could help treat cancer, especially when DNA methyltransferase inhibitors (like azacytidine) and histone deacetylase inhibitors (like vorinostat) are used. In clinical studies, they have shown that they can reactivate tumor suppressor genes and restore normal gene expression. This can cause tumors to shrink and patients with blood cancers to have better results. But problems like drug tolerance, harm, and different reactions from patients are still big problems. Targeting non-coding RNAs and mixing epigenetic medicines with immunotherapy or chemotherapy has been shown to work better together, making treatments more effective. As we learn more about CRISPR-based histone editing and AI-driven drug research, these medicines will likely be improved even more, making cancer care more personalized and exact.
|
Table 2. Clinical Efficacy of Epigenetic Inhibitors (DNMTis, HDACis) in Cancer Treatment |
|||
|
Cancer Type |
Response Rate (%) |
Progression-Free Survival (Months) |
Overall Survival (Months) |
|
Myelodysplastic Syndromes |
45 |
16 |
28 |
|
Cutaneous T-Cell Lymphoma |
55 |
12 |
22 |
|
Acute Myeloid Leukemia |
38 |
15 |
24 |
|
Cutaneous T-Cell Lymphoma |
50 |
18 |
26 |
The information shown shows how well epigenetic inhibitors work in treating different types of cancer, including Myelodysplastic Syndromes (MDS), Cutaneous T-Cell Lymphoma (CTCL), and Acute Myeloid Leukemia (AML). Response rates and life rates show how well drugs like DNA methyltransferase inhibitors (DNMTis) and histone deacetylase inhibitors (HDACis) work in treating these types of cancer.
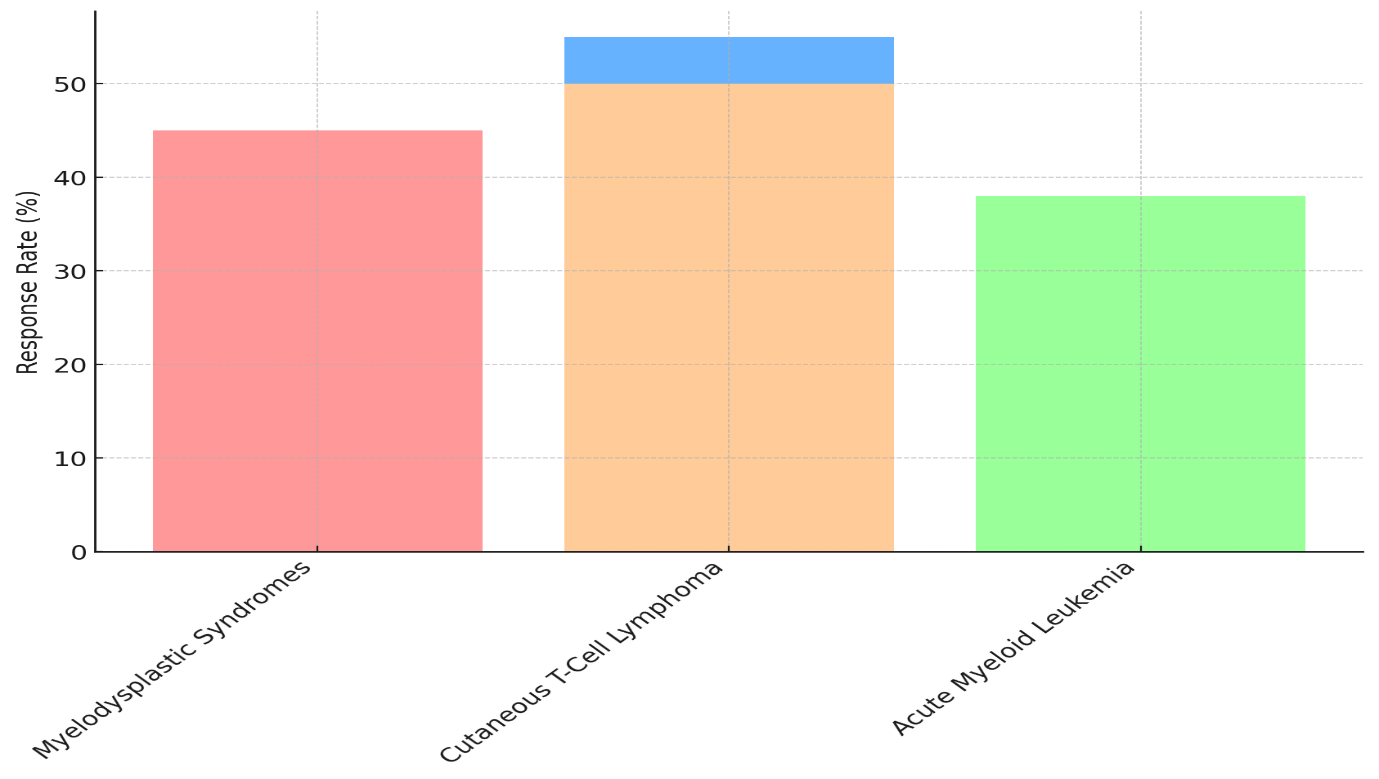
Figure 3. Response Rates Across Hematological Conditions
A response rate of 45 % and a total mortality rate of 28 months for Myelodysplastic Syndromes show that the treatment is somewhat effective. The 16-month progression-free survival rate shows that the disease can be controlled for a long time, but relapses may happen soon after the first treatment. The responder rate is a little higher in Cutaneous T-Cell Lymphoma (55 %), but the progression-free survival is only 12 months, and the total survival is 22 months. This shows in figure 3 that even though the medicine works, CTCL is still hard to deal with because the recovery times are shorter.
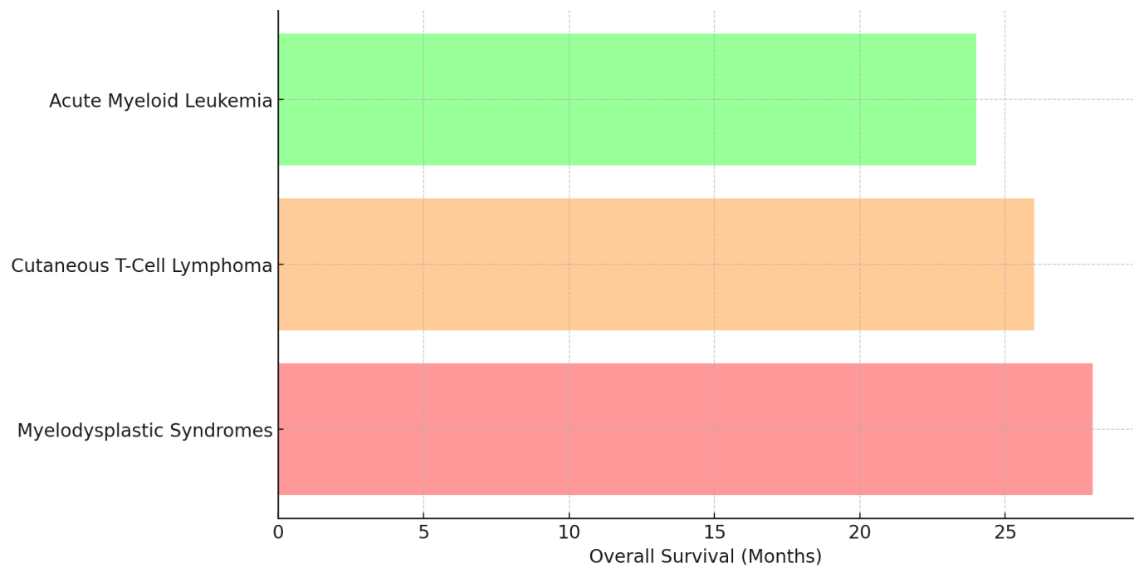
Figure 4. Overall Survival Comparison Among Hematological Conditions
For Acute Myeloid Leukemia, the reaction rate is only 38 %, and the total survival rate is 24 months. The progression-free survival rate is 15 months. Because AML is a more severe type of cancer, these epigenetic medicines may not work as well on it, as shown by the lower reaction rates and survival rates, shown in figure 4.
|
Table 3. Combination Therapies with Epigenetic Modifiers |
||||
|
Combination Therapy |
Cancer Type |
Response Rate (%) |
Overall Survival (Months) |
Adverse Effects (%) |
|
Azacytidine + Chemotherapy |
Myelodysplastic Syndromes |
60 |
30 |
25 |
|
Vorinostat + Chemotherapy |
Non-Hodgkin Lymphoma |
65 |
28 |
30 |
|
Azacytidine + Immunotherapy |
Acute Myeloid Leukemia |
50 |
26 |
20 |
|
Romidepsin + Immunotherapy |
Cutaneous T-Cell Lymphoma |
70 |
32 |
35 |
When used together, Azacytidine and chemotherapy have a 60 % response rate and a total mortality rate of 30 months for people with Myelodysplastic Syndromes (MDS). However, 25 % of patients experience side effects. This shows that using both epigenetic treatment and chemotherapy together can help treat MDS, leading to a higher chance of mortality compared to using Azacytidine alone while keeping side effects to a reasonable level. Vorinostat plus chemotherapy 65 % of people with non-Hodgkin lymphoma get better and live for 28 months, with 30 % having bad effects, represent in figure 5. The mixture seems to work better than chemotherapy by itself, but it also has a few more side effects, which is a problem that often comes up with combination treatments.
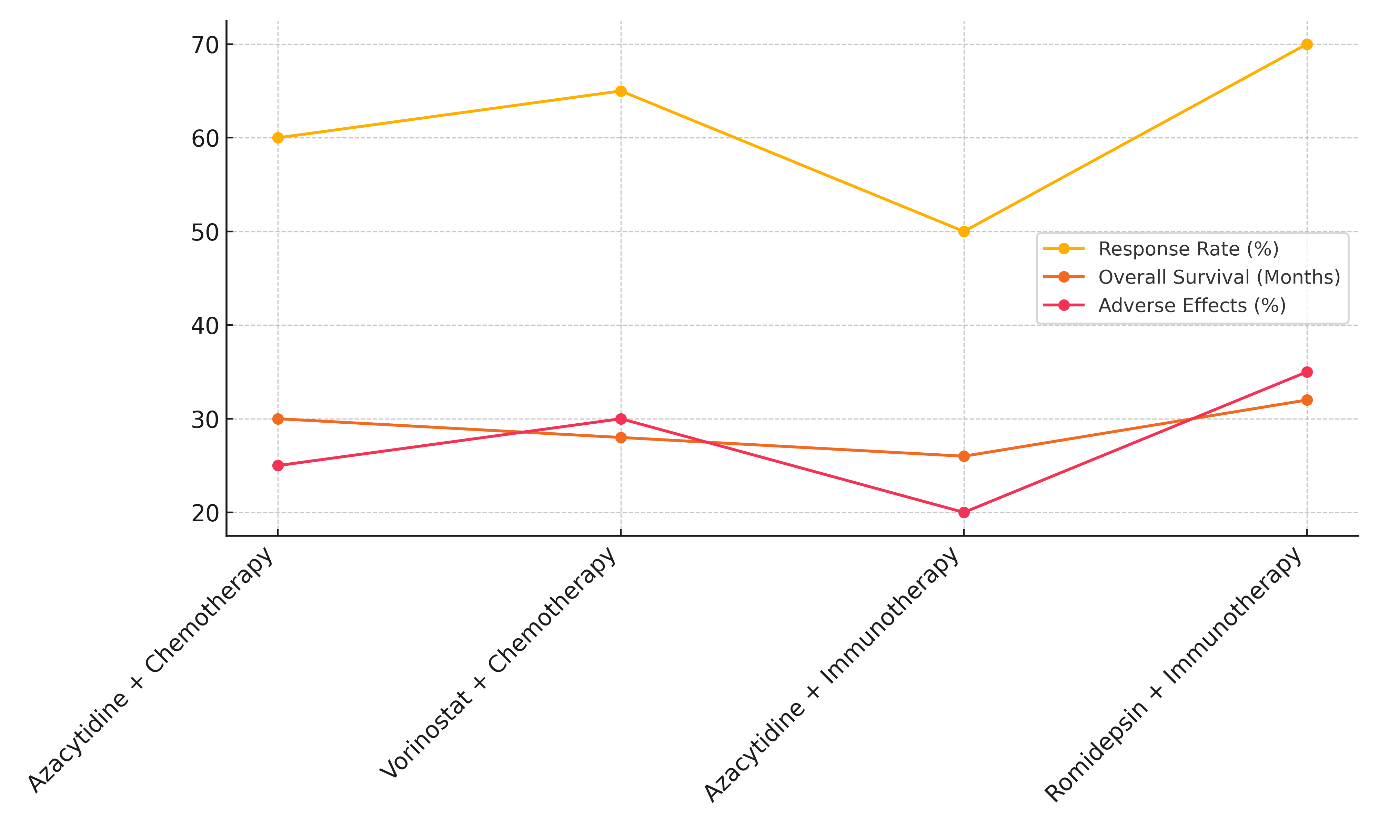
Figure 5. Therapy Effectiveness: Response Rate, Survival, and Adverse Effects
In Acute Myeloid Leukemia (AML), Azacytidine plus immunotherapy has a 50 % response rate and a total survival rate of 26 % after 26 months. It also has a reasonably low rate of side effects (20 %), illustrate in figure 6
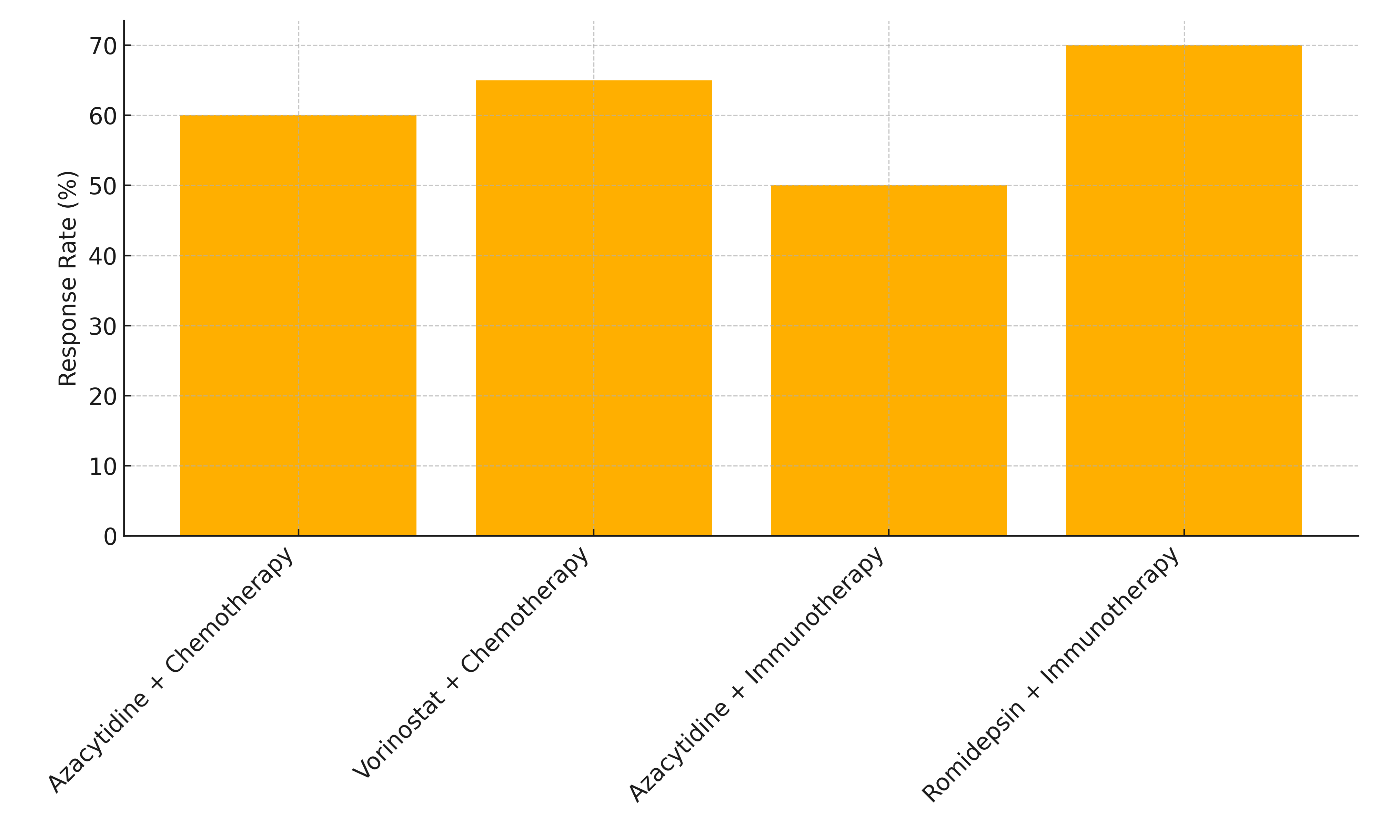
Figure 6. Comparison of Response Rates by Therapy Type
Even though this mixture seems to help people survive, the reaction rate is still pretty low. This suggests that more testing or combination methods may be needed to make it more effective in AML. In the end, Romidepsin plus immunotherapy has the best reaction rate (70 %), with a 32-month total survival rate in Cutaneous T-Cell Lymphoma, though it also has the worst side effects (35 %). This shows that even though this mixture works very well, the most important thing to do to improve patient results is to control the side effects.
CONCLUSIONS
Epigenetic changes have become very important in controlling gene expression and the growth of cancer tumors. New epigenetic treatments have been created as scientists learn more about how abnormal DNA methylation, histone changes, and non-coding RNA instability help cancer spread. DNA methyltransferase inhibitors (DNMTis) and histone deacetylase inhibitors (HDACis) have been shown to be effective in the treatment of cancer, especially blood cancers, by turning on tumor suppressor genes that have been turned off by epigenetics and improving the reaction to standard treatments when used together. Besides that, focusing on non-coding RNAs like microRNAs and long non-coding RNAs opens up new ways to change gene expression and stop tumor growth. Although the results look good, there are still some problems that need to be fixed before epigenetic treatments can be used in people. To get around the big problems of drug tolerance, side effects, and harm, we need better ways to give drugs and use them together in treatments. New inventions, like CRISPR-based epigenetic editing, may help by letting exact changes be made to the epigenome. This could fix the basic reasons why cancer cells have epigenetic changes. Additionally, artificial intelligence (AI) is becoming more and more important in finding new epigenetic targets, improving drug research, and tailoring cancer treatment plans for each patient. The future of epigenetic cancer treatment lies in learning more about the biological processes that control the epigenome and making medicines that are more focused, effective, and less harmful. Personalized medicine, which is based on AI-driven insights and DNA profiles, could lead to more effective and individualized treatment plans that help patients do better and lower their risk of return. In conclusion, epigenetic therapy is still a new area that is growing. However, it is already a game-changing way to treat cancer, giving patients who haven't had many treatment choices new hope and moving precision oncology forward.
BIBLIOGRAPHIC REFERENCES
1. Li, M.; Zhang, D. DNA methyltransferase-1 in acute myeloid leukaemia: Beyond the maintenance of DNA methylation. Ann. Med. 2022, 54, 2011–2023.
2. Fiñana, C.; Gómez-Molina, N.; Alonso-Moreno, S.; Belver, L. Genomic and Epigenomic Landscape of Juvenile Myelomonocytic Leukemia. Cancers 2022, 14, 1335.
3. Li, J.; Li, L.; Wang, Y.; Huang, G.; Li, X.; Xie, Z.; Zhou, Z. Insights into the Role of DNA Methylation in Immune Cell Development and Autoimmune Disease. Front. Cell Dev. Biol. 2021, 9, 757318.
4. Sharma, V.; Wright, K.L.; Epling-Burnette, P.K.; Reuther, G.W. Metabolic Vulnerabilities and Epigenetic Dysregulation in Myeloproliferative Neoplasms. Front. Immunol. 2020, 11, 604142.
5. Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183.
6. Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651.
7. Ho, P.T.B.; Clark, I.M.; Le, L.T.T. MicroRNA-Based Diagnosis and Therapy. Int. J. Mol. Sci. 2022, 23, 7167
8. Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug. Discov. 2017, 16, 203–222.
9. Jones, P.A.; Issa, J.P.J.; Baylin, S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016, 17, 630–641.
10. Daher-Reyes, G.S.; Merchan, B.M.; Yee, K.W.L. Guadecitabine (SGI-110): An investigational drug for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Expert Opin. Investig. Drugs. 2019, 28, 835–849.
11. Wen, T.; Sun, G.; Jiang, W.; He, X.; Shi, Y.; Ma, F.; Liu, P. Histone deacetylases inhibitor chidamide synergizes with humanized PD1 antibody to enhance T-cell chemokine expression and augment Ifn-γ response in NK-T cell lymphoma. eBioMedicine 2023, 87, 104420.
12. Adair, J.E.; Johnston, S.K.; Mrugala, M.M.; Beard, B.C.; Guyman, L.A.; Baldock, A.L.; Bridge, C.A.; Hawkins-Daarud, A.; Gori, J.L.; Born, D.E.; et al. Gene therapy enhances chemotherapy tolerance and efficacy in glioblastoma patients. J. Clin. Investig. 2014, 124, 4082–4092.
13. Ruan, J.; Moskowitz, A.J.; Mehta-Shah, N.; Sokol, L.; Chen, Z.; Rahim, R.; Song, W.; Van Besien, K.; Horwitz, S.M.; Rutherford, S.C. Multi-Center Phase II Study of Oral Azacitidine (CC-486) Plus CHOP As Initial Treatment for Peripheral T-Cell Lymphoma (PTCL). Blood 2020, 136 (Suppl. S1), 33–34.
14. Montesinos, P.P.; Roboz, G.J.; Bulabois, C.E.; Subklewe, M.; Platzbecker, U.; Ofran, Y.; Papayannidis, C.; Wierzbowska, A.; Shin, H.J.; Doronin, V. Safety and efficacy of talacotuzumab plus decitabine or decitabine alone in patients with acute myeloid leukemia not eligible for chemotherapy: Results from a multicenter, randomized, phase 2/3 study. Leukemia 2021, 35, 62–74.
15. R. Golchha, P. Khobragade and A. Talekar, “Design of an Efficient Model for Health Status Prediction Using LSTM, Transformer, and Bayesian Neural Networks,” 2024 International Conference on Innovations and Challenges in Emerging Technologies (ICICET), Nagpur, India, 2024, pp. 1-5, doi: 10.1109/ICICET59348.2024.10616353.
16. Sébert, M.; Renneville, A.; Bally, C.; Peterlin, P.; Beyne-Rauzy, O.; Legros, L.; Gourin, M.P.; Sanhes, L.; Wattel, E.; Gyan, E.; et al. A phase II study of guadecitabine in higher-risk myelodysplastic syndrome and low blast count acute myeloid leukemia after azacitidine failure. Haematologica 2019, 104, 1565–1571.
17. Salamero, O.; Montesinos, P.; Willekens, C.; Pérez-Simón, J.A.; Pigneux, A.; Récher, C.; Popat, R.; Carpio, C.; Molinero, C.; Mascaró, C.; et al. First-in-Human Phase I Study of Iadademstat (ORY-1001): A First-in-Class Lysine-Specific Histone Demethylase 1A Inhibitor, in Relapsed or Refractory Acute Myeloid Leukemia. J. Clin. Oncol. 2020, 38, 4260–4273.
18. Navarro Mendivil, A.F.; Gutierrez, S.; Bullock, R.; Buesa, C. 1806P Final safety and efficacy data from CLEPSIDRA trial in 2L ED-SCLC. Ann. Oncol. 2020, 31, S1044.
19. Kurmasheva, R.T.; Erickson, S.W.; Han, R.; Teicher, B.A.; Smith, M.A.; Roth, M.; Gorlick, R.; Houghton, P.J. In vivo evaluation of the lysine-specific demethylase (KDM1A/LSD1) inhibitor SP-2577 (Seclidemstat) against pediatric sarcoma preclinical models: A report from the Pediatric Preclinical Testing Consortium (PPTC). Pediatr. Blood Cancer 2021, 68, e29304.
20. Lin, Y.C.; Lin, J.H.; Chou, C.W.; Chang, Y.F.; Yeh, S.H.; Chen, C.C. Statins increase p21 through inhibition of histone deacetylase activity and release of promoter-associated HDAC1/2. Cancer Res. 2008, 68, 2375–2383.
21. Bridgeman, S.; Northrop, W.; Ellison, G.; Sabapathy, T.; Melton, P.E.; Newsholme, P.; Mamotte, C.D.S. Statins Do Not Directly Inhibit the Activity of Major Epigenetic Modifying Enzymes. Cancers 2019, 11, 516.
22. Lauria, A.; Mannino, S.; Gentile, C.; Mannino, G.; Martorana, A.; Peri, D. DRUDIT: Web-based DRUgs DIscovery Tools to design small molecules as modulators of biological targets. Bioinformatics 2020, 36, 1562–1569.
FINANCING
None.
CONFLICT OF INTEREST
Authors declare that there is no conflict of interest.
AUTHORSHIP CONTRIBUTION
Conceptualization: Kukatla Tejesh, Lovish Dhingra, Vinima Gambhir, Amit Kumar, Nibedita Sahoo, Manti Debnath, Malathi. H.
Data curation: Kukatla Tejesh, Lovish Dhingra, Vinima Gambhir, Amit Kumar, Nibedita Sahoo, Manti Debnath, Malathi. H.
Formal analysis: Kukatla Tejesh, Lovish Dhingra, Vinima Gambhir, Amit Kumar, Nibedita Sahoo, Manti Debnath, Malathi. H.
Drafting - original draft: Kukatla Tejesh, Lovish Dhingra, Vinima Gambhir, Amit Kumar, Nibedita Sahoo, Manti Debnath, Malathi. H.
Writing - proofreading and editing: Kukatla Tejesh, Lovish Dhingra, Vinima Gambhir, Amit Kumar, Nibedita Sahoo, Manti Debnath, Malathi. H.